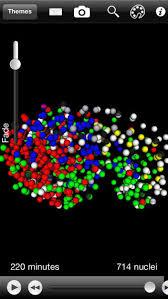
WormGUIDES Atlas
WormGUIDES Atlas is an interactive 4D portrayal of neural development in C. elegans. It will ultimately contain nuclear positions for every cell in the embryo, identified and tracked from the 2 cell stage until hatching. Single-cell and subcellular information, including neural outgrowth dynamics for each cell as well as cell function, gene expression, the adult neural connectome and related literature will be collated for each cell from public sources and also integrated with the atlas model. WormGUIDES Atlas integrates tools for exploratory data analyses and insight sharing. Navigation is linked between 3D and lineage tree views. In both contexts, community single cell information can be accessed with a click, creating live web queries that summarize knowledge about a cell. In many cases this information can be used to control cell color, creating customized interactive visualizations. A user's insights can be annotated directly into the embryo model with a note-taking interface that attaches each annotation to a cell or other point in space and time. These multi-dimensionally located notes can then be ordered into a (chrono)logical story sequence that explains developmental events as they unfold in the embryo. Annotations can be saved and shared with collaborators or the community.