Description
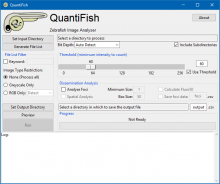
The software FishInspector provides automatic feature detections in images of zebrafish embryos (body size, eye size, pigmentation). It is Matlab-based and provided as a Windows executable (no matlab installation needed).
The recent version requires images of a lateral position. It is important that the position is precise since deviation may confound with feature annotations. Images from any source can be used. However, depending on the image properties parameters may have to be adjusted. Furthermore, images obtained with normal microscope and not using an automated position system with embryos in glass capillaries require conversion using a KNIME workflow (the workflow is available as well). As a result of the analysis the software provides JSON files that contain the coordinates of the features. Coordinates are provided for eye, fish contour, notochord , otoliths, yolk sac, pericard and swimbladder. Furthermore, pigment cells in the notochord area are detected. Additional features can be manually annotated. It is the aim of the software to provide the coordinates, which may then be analysed subsequently to identify and quantify changes in the morphology of zebrafish embryos.