Description
SOAX is an open source software tool to extract the centerlines, junctions and filament lengths of biopolymer networks in 2D and 3D images. It facilitates quantitative, reproducible and objective analysis of the image data. The underlying method of SOAX uses multiple Stretching Open Active Contours (SOACs) that are automatically initialized at image intensity ridges and then stretch along the centerlines of filaments in the network. SOACs can merge, stop at junctions, and reconfigure with others to allow smooth crossing at junctions of filaments.
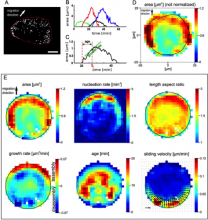
SOAX provides 3D visualization for exploring image data and visually checking results against the image. Quantitative analysis functions based on extracted networks are also implemented in SOAX, including spatial distribution, orientation, and curvature of filamentous structures. SOAX also provides interactive manual editing to further improve the extraction results, which can be saved in a file for archiving or further analysis. Useful for microtubules or actin filaments.
Observation: Depending on the operating system, the installation may or may not require Boost C++, ITK and VTK libraries. Windows has a standalone executable application without the need of those.