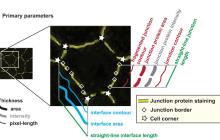
Segmentation of membrane of mouse, sea urchin and human oocytes from transmitted light images
This dataset contains images acquired in transmitted light with different settings of mouse and human oocytes and sea urchin eggs, with the corresponding ground-truth of the membrane segmentation. Mouse oocyte images were taken before and during oocyte maturation (meiosis I). Some human oocyte images were taken during oocyte maturation (meiosis I), and some are M-II oocytes just after fertilization. Sea urchin images contains both fertilized and unfertilized eggs.